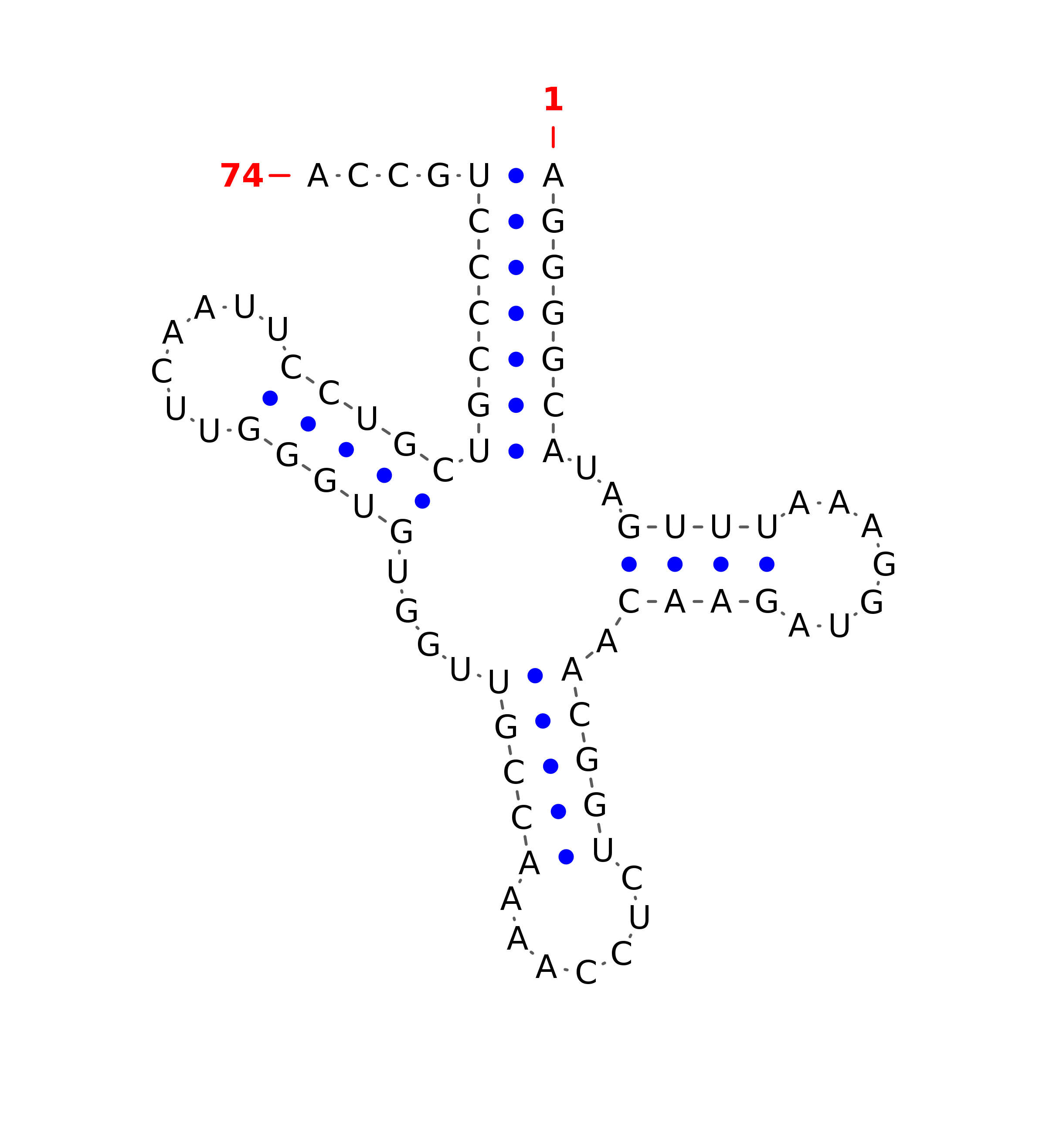
T-box riboswitch DTHUZBQL
Putative MET (CAU) T-box riboswitch from Enterococcus saccharolyticus subsp. saccharolyticus ATCC 43076
Source information
| TBDB ID | DTHUZBQL |
| Host organism | Enterococcus saccharolyticus subsp. saccharolyticus ATCC 43076 |
| Model used | Class I |
| Predicted regulation | Unknown |
| T-box riboswitch genomic accession | KE136389.1:379462-379747 |
| Downstream protein description | hypothetical protein |
| Downstream protein ID | GenBank: EOT30669.1 |
| T-box riboswitch length | |
| Specifier loop sequence | AAUGG |
| T-box 5'-UGGN-3' sequence | UGGU |
| Predicted specifier | AUG |
| Predicted tRNA family | MET (CAU) |
| Found cognate tRNA | True |
| Alternative specifiers | AAU,UGG |
| Metagenome | False |
| Structural predictions | Partial |
Genomic Context
Error loading NCBI Genome Browser - Cookies disabled
To use the NCBI genome browser for visualizing genetic context of T-box riboswitch sequences, please enable cookies on your web browser.
Full T-box riboswitch FASTA sequence
>T-box Riboswitch Sequence DTHUZBQL
TAAAATTCAATGAAGAGAAAAGTACACTTCGCAAATTTTAAGAGAGGTTCTGGATGGTGCAAAGAACTAATTTAATAAGTAGAAAAATGGTCTCTGAGAAAAACTGTTGATAAGTAAGCAGTTTCGGAAAAGTTCATCCGTTATATTTGAACGCCAATCTCATGATAGATGATGTTGGACAAAGAGAAGTGCAAGCTTCTAAACAAAGGTGGTACCGCGATATATTCGCCCTTTTACTGGTTTTTTTAACTGGTAAAAGGGCTTTTTTT
Predicted secondary structures
T-box riboswitch DTHUZBQL Antiterminator
......((......((((......(((((.....(((........((((((...........))))))))).....)))).)........))))...))..(((((((........))))))).(((..........)))............................................((((((....)))))).....((((.......((((.....))))))))....................................
T-box riboswitch DTHUZBQL Terminator
Error: TBDB structure refinement failed to predict a terminator structure for this T-box
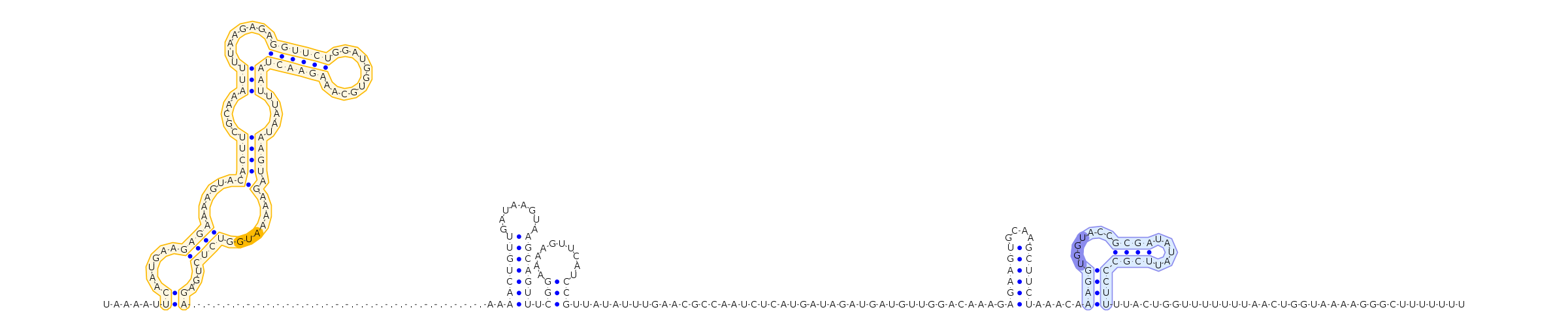

Specifier and tRNA predictions
Top specifier
Top predicted specifier: AUGTop predicted tRNA family: MET (CAU) Best tRNA match for top specifier >MET (CAU) tRNA from Enterococcus saccharolyticus subsp. saccharolyticus ATCC 43076
CGCGGTGTAGCTCAGCTGGCTAGAGCGTCCGGTTCATACCCGGGAGGTCGGGGGTTCGATCCCCTCCGCCGCGACCA
Predicted tRNA structure (((((((..((((.........)))).(((((.......))))).....(((((.......))))))))))))....
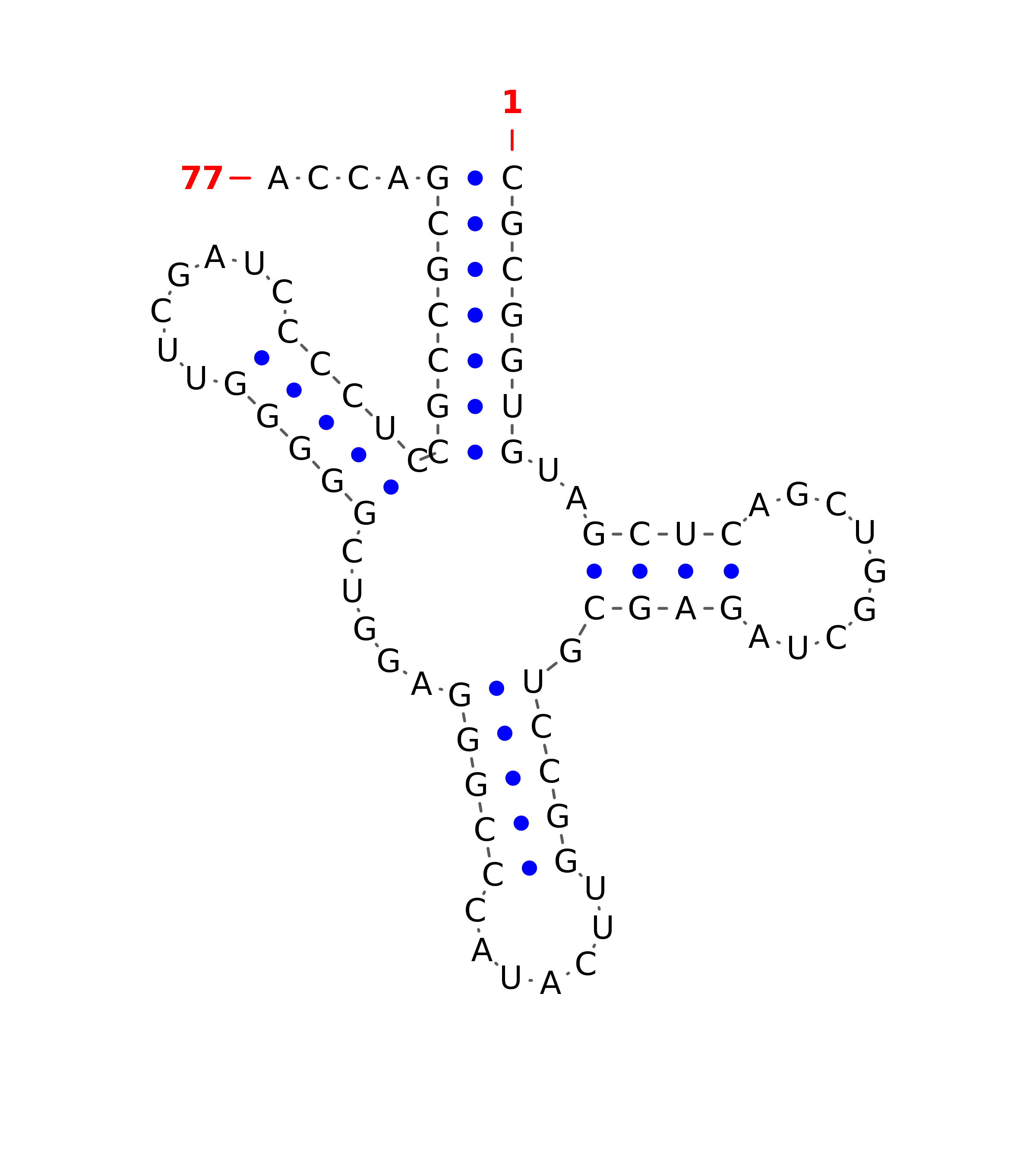
Minimum Free Energy (MFE) Calculations
INFERNAL T-box riboswitch prediction output
| E-Value | |
| Score | 117.9 |
| Bias | 4.3 |
| CM Accuracy | 0.94 |
*More information about the meaning of these values can be found here.
| Feature | Length | Locus |
| Stem I | [7,99] | |
| Stem II region | ||
| Stem III | ||
| Specifier region | - | [86,88] |
| T-box UGGN locus | - | [210,213] |
| Antiterminator | [206,233] | |
| Terminator | [228,269] | |
| All stems | - | [[7, 99], [102, 123], [125, 140], [185, 200], [206, 233]] |
Input source sequence
>KE136389.1:379462-379747
TAAAATTCAATGAAGAGAAAAGTACACTTCGCAAATTTTAAGAGAGGTTCTGGATGGTGCAAAGAACTAATTTAATAAGTAGAAAAATGGTCTCTGAGAAAAACTGTTGATAAGTAAGCAGTTTCGGAAAAGTTCATCCGTTATATTTGAACGCCAATCTCATGATAGATGATGTTGGACAAAGAGAAGTGCAAGCTTCTAAACAAAGGTGGTACCGCGATATATTCGCCCTTTTACTGGTTTTTTTAACTGGTAAAAGGGCTTTTTTT
TAAAATTCAATGAAGAGAAAAGTACACTTCGCAAATTTTAAGAGAGGTTCTGGATGGTGCAAAGAACTAATTTAATAAGTAGAAAAATGGTCTCTGAGAAAAACTGTTGATAAGTAAGCAGTTTCGGAAAAGTTCATCCGTTATATTTGAACGCCAATCTCATGATAGATGATGTTGGACAAAGAGAAGTGCAAGCTTCTAAACAAAGGTGGTACCGCGATATATTCGCCCTTTTACTGGTTTTTTTAACTGGTAAAAGGGCTTTTTTT
INFERNAL output sequence
>Infernal-KE136389.1:379462-379747
UAAAAUUCAAUGAAGAGA-AAAGUAC-ACUUCG--CAAAUUUUAAGAGAGGUUCUGGAUGGUGCAAAGAACUAAUUUAAUAAGUAGAAAAAUGGUCUCUGAGAAAAACUGUUGAU-----AAGUAAGCAGUUUCGGAAAAguucAUCCGUUAUauuugaacgccaaucucaugauagaUGAUGUUGGACAAAGAGAAGUG------CAAGCUUCUAAAcAAAGGUGGUACCGCGAU--AUA-UUCGCCCUUUUA
UAAAAUUCAAUGAAGAGA-AAAGUAC-ACUUCG--CAAAUUUUAAGAGAGGUUCUGGAUGGUGCAAAGAACUAAUUUAAUAAGUAGAAAAAUGGUCUCUGAGAAAAACUGUUGAU-----AAGUAAGCAGUUUCGGAAAAguucAUCCGUUAUauuugaacgccaaucucaugauagaUGAUGUUGGACAAAGAGAAGUG------CAAGCUUCUAAAcAAAGGUGGUACCGCGAU--AUA-UUCGCCCUUUUA
INFERNAL output structure - (Antiterminator)
>Infernal-Structure-KE136389.1:379462-379747
::::::<<------<<<<-------<<<<<<-------<<<--------<<<<<<___________>>>>>>>>>----->>>>>>-------->>>>--->>,,<<<<<<<_____________>>>>>>>,<<<____....__>>>,,,,.........................,,,,,,,,,,,,,,,<<<<<<<________>>>>>>>,,,.,<<<<-------<<<<<______>>>>>>>>>:::
::::::<<------<<<<-------<<<<<<-------<<<--------<<<<<<___________>>>>>>>>>----->>>>>>-------->>>>--->>,,<<<<<<<_____________>>>>>>>,<<<____....__>>>,,,,.........................,,,,,,,,,,,,,,,<<<<<<<________>>>>>>>,,,.,<<<<-------<<<<<______>>>>>>>>>:::