What is the T-box riboswitch leader sequence?
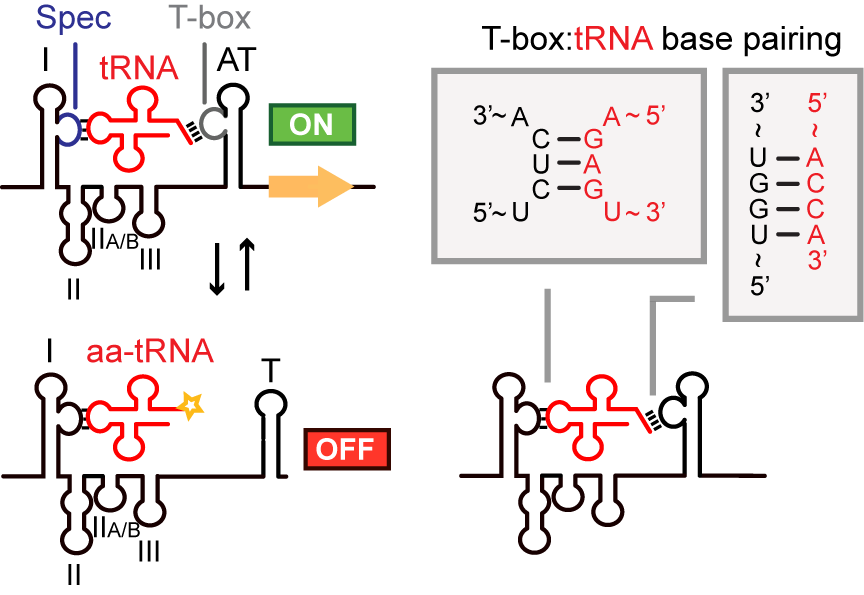
The T-box riboswitch leader is a bacterial RNA regulatory element found upstream of many gene clusters associated with amino acid biosynthesis and tRNA-aminoacylation. T-box riboswitches are capable of sensing tRNA aminoacylation state. When an uncharged tRNA is bound, an antiterminator hairpin is formed; otherwise, a terminator hairpin is formed. Downstream genes are therefore transcribed only in the presence of uncharged tRNA. For more information, see the Rfam entry on T-box riboswitches .
T-box riboswitch features
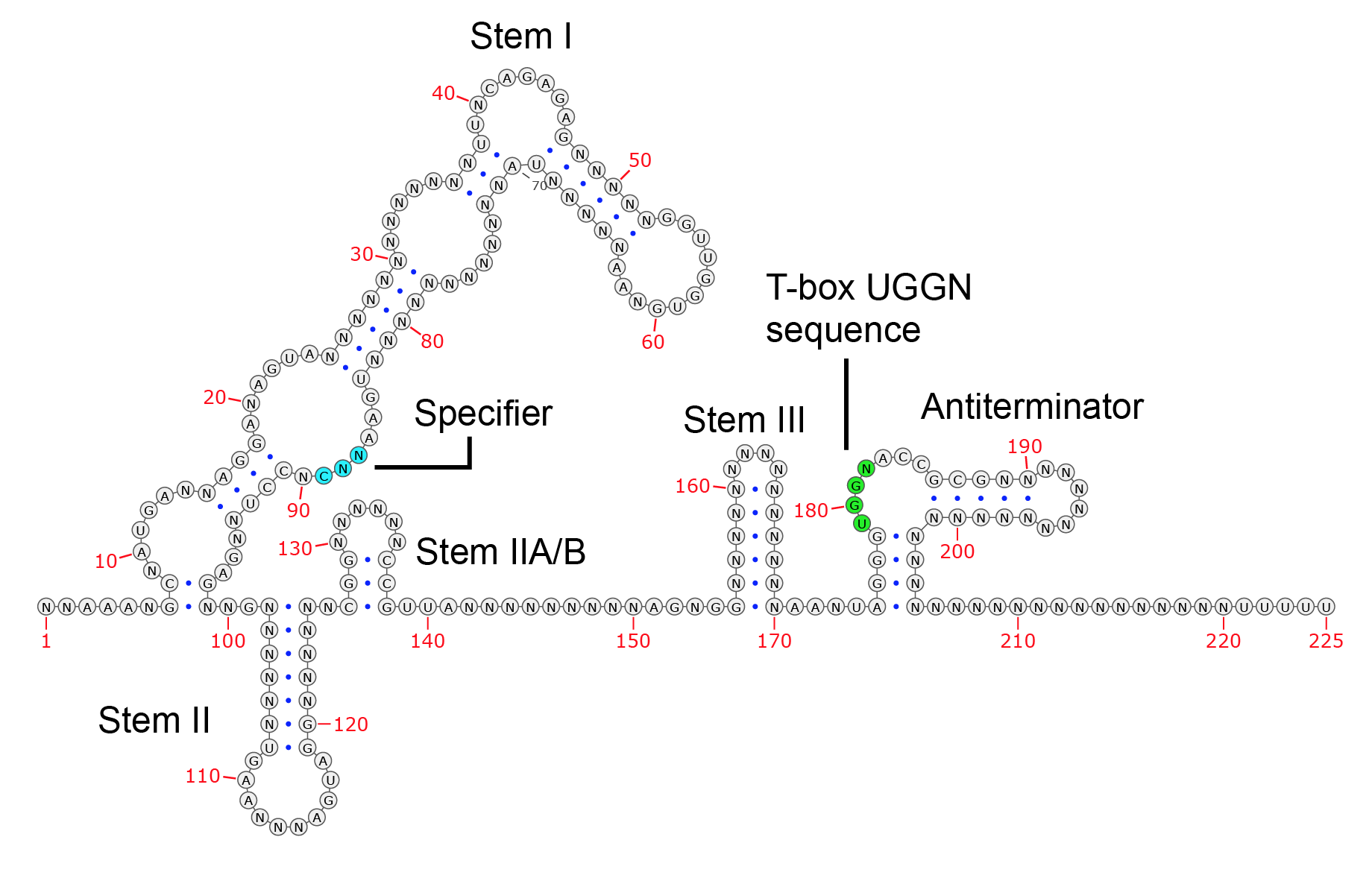
T-box riboswitches have the following structural features. The image below shows the consensus structures of T-box riboswitches within the TBDB. Note that structures can vary from the conserved set.
- Stem I: One of the first stems in a T-box riboswitch. Has several loops, and contacts the tRNA anticodon at the specifier. In certain T-box riboswitches, Stem I also makes contact with the tRNA D-loop.
- Specifier loop: Within Stem I, the specifier loop pairs with the anticodon of the cognate tRNA. This helps determine the T-box riboswitch specificity. May contain multiple specifier frames capable of binding more than one tRNA species.
- Stem II: T-box riboswitches often have additional Stem II structures following the stem I, and in rare cases preceding stem I.
- Stem IIA/B: This pseudoknot structure is present on T-box riboswitches directly after the Stem II (if present), though exceptions exist.
- Stem III: Structural loop that precedes the antiterminator/antisequestrator in T-box riboswitches .
- Anti-terminator/antisequestrator: The antiterminator (transcriptional T-box riboswitches ) or antisequestrator (translational T-box riboswitches ) is a hairpin near the 3’ end of the T-box, containing a bulge with a 5'-UGGN-3' sequence that pairs with the 5'-NCCA-3' of an uncharged tRNA.
T-box riboswitches in synthetic biology
T-box riboswitches can be useful tools in synthetic biology. In accordance with their natural function, they can be used to direct transcription of downstream genes based on tRNA aminoacylation state.
More broadly, they can also be used as tRNA-binding elements. For example, Ishida et al. showed that a T-box riboswitch can be fused to an aminoacylating ribozyme to confer specificity to tRNAs recognized by the T-box riboswitch.
Predicting downstream gene function based on T-box riboswitch specificity
Knowing the specificity of a T-box riboswitch regulating a downstream gene may be helpful in predicting that gene's function. For example, amino acid transporters are often difficult to classify, but transporters regulated by T-box riboswitches can be predicted to transport the same amino acid that the T-box riboswitch senses.
T-box riboswitches as antibiotic targets
Since T-box riboswitches are found only in bacteria, they may be attractive antibiotic targets. For example, Frolich et al. demonstrated that small-molecule inhibitors of T-box/tRNA binding can inhibit the growth of Gram-positive bacteria in vitro.
Key references
- Initial T-box riboswitch annotation (n=698), and transporter specificity prediction: Vitreschak et al., 2008
- Using covariance models with INFERNAL to detect RNA motifs and predict secondary structure: Nawrocki and Eddy, 2013
- The Rfam database: Kalvari et al., 2018
- T-box riboswitch Stem I crystal structure and interactions with tRNA: Grigg et al., 2013, and Zhang and D'Amare, 2013
- T-box riboswitch cryo-EM structure, and mechanism of aminoacylation sensing: Li et al., 2019
- T-box riboswitch crystal structure, and mechanism of aminoacylation sensing: Battaglia et al., 2019
- ileS translational T-box riboswitch crystal structure: Suddala and Zhang, 2019

